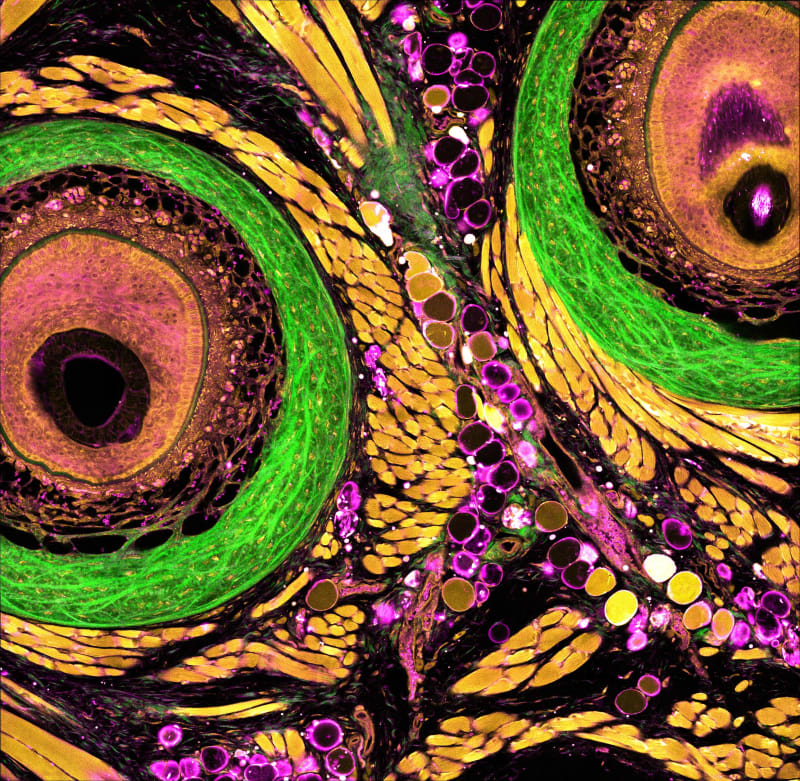
Understanding how cells behave and change within living tissues is essential for advancing medicine-whether it's studying how tumors grow, testing the effectiveness of new drugs, or exploring how the immune system responds to infection and injury.
Our team has created a new powerful imaging platform called dSLAM (deep and dynamic simultaneous label-free imaging of autofluorescence and multiharmonic). This system enables researchers to visualize live, intact tissues in action-without the need for any fluorescent dyes or genetic labels, with intrinsic metabolic and structural contrast, at three times deeper depth than existing label-free imaging platforms. Label-free imaging uses the cells' natural glow, known as autofluorescence, to monitor metabolic activity in a completely noninvasive way. While this concept has long held promise, traditional approaches have struggled with poor resolution, limited imaging depth, and slow data collection. We addressed these challenges with a set of novel innovations:
- Sharper, deeper label-free imaging: By using a three-photon NADH excitation, we significantly reduce how much light scatters inside tissue. As a result, we can now capture clear metabolic images more than 700 microns below the tissue surface-over twice the depth possible with older technologies-without sacrificing resolution.
- Custom laser light: To make this possible, we engineered a new kind of light delivery system that uses shaped multimode optical fibers. This design produces intense but finely controlled bursts of light at desired new wavelengths (1110 nm, up to 0.5 megawatts in peak power) while avoiding damage to sensitive biological samples.
- AI-powered clarity: We also developed an artificial intelligence tool called SSAI-3D, a self-supervised software system that removes blur from deep tissue images. This approach works across various microscope types and sample conditions, and it doesn't require any special hardware or labeled training data. With SSAI-3D, we recover full 3D resolution-around 500 nanometers in the three xyz dimensions-even in thick, complex tissue.
Together, these technologies make it possible to observe the structural and metabolic changes of living systems with unmatched depth, speed, and precision. Importantly, the platform is adaptable across different types of samples and imaging setups.
We've already used dSLAM to solve pressing biomedical problems. In collaboration with MIT's Prof. Roger Kamm, we imaged a full 700-micron depth of a human blood-brain barrier model, helping to visualize how therapeutic drugs may (or may not) pass into the brain. In a separate project with MIT’s Prof. Linda Griffith, we captured dynamic changes in human endometrial tissue across the menstrual cycle. This research is opening new doors in understanding hormone response, treatment effects, and patient-specific differences in reproductive health.
Our work has been published in Science Advances and Nature Communications. The technology has also been featured in multiple major media outlets, including Tech Briefs. Our long-term goal is to continue applying this platform to tackle critical medical challenges-and to further empower deep metabolic imaging technology through new optical phenomena and algorithms, creating a cycle of solution innovation driven by real-world needs.
Video
Like this entry?
-
About the Entrant
- Name:Kunzan Liu
- Type of entry:teamTeam members:
- Honghao Cao
- Linda Griffith
- Sixian You
- Software used for this entry:Custom written Labview control and python processing
- Patent status:pending